1 Introduction
Template strand/antisense strand/minus strand: 在转录时,RNA 聚合酶以该链为模板,依据碱基互补配对原则合成 mRNA。简言之,此条链的碱基与 mRNA 的碱基互补配对。
Coding strand/sense strand/plus strand: 序列与 mRNA 相同(仅 T 变为 U)。
Transcription can occur in both directions, genes being located on either DNA strand, sometimes overlapping. In eukaryotes, a complementary RNA molecule to a given mRNA can also be transcribed: this has been described as antisense transcription and these molecules are involved in regulatory mechanism. Knowing from which DNA strand the RNA molecule originates from is an important piece of information, which helps resolving annotation ambiguities for known and novel genes, provides hints to the function of the studied RNA, and helps to correctly predict the expression levels of a given transcript.
While in non-strand specific RNA-Seq, reads corresponding to RNA molecule or complementary to it are indistinguishable. Strand-specific RNA-Seq can achieve this goal.
At present, commonly used strand-specific RNA-Seq protocols are mainly derived from the following two principles:
- Via direct ligation of adaptors to RNA molecules: there are no restrictions in the RNA length and it is the only choice for the analysis of short RNA molecules like micro-RNA, but it is sensitive to rRNA cantamination, so the RNA fraction of interest must be preselected.
Why?
This method directly attaches adaptors to the 3’ ends of all RNA molecules regardless of their types, so it has no restrictions in the RNA length and in RNA types.
ds cDNA synthesis-derived methods reply on the poly-A tail, short RNAs do not have poly-A tails.
rRNA constitues > 80~90% of total RNA in a typical cell, which means that it massively outnumbers the RNA of interest, so preselection step is essential.
Random priming used to synthesize the 1st cDNA strand repuires a template long enough for a random primer to bind stably.
Specific priming requires a known sequence to design a primer, which isn’t feasible for diverse unknown short RNAs.
TSO-dependent cDNA synthesis also needs a minimum RNA length for the reverse transcriptase enzyme to work effectively and add the TSO.
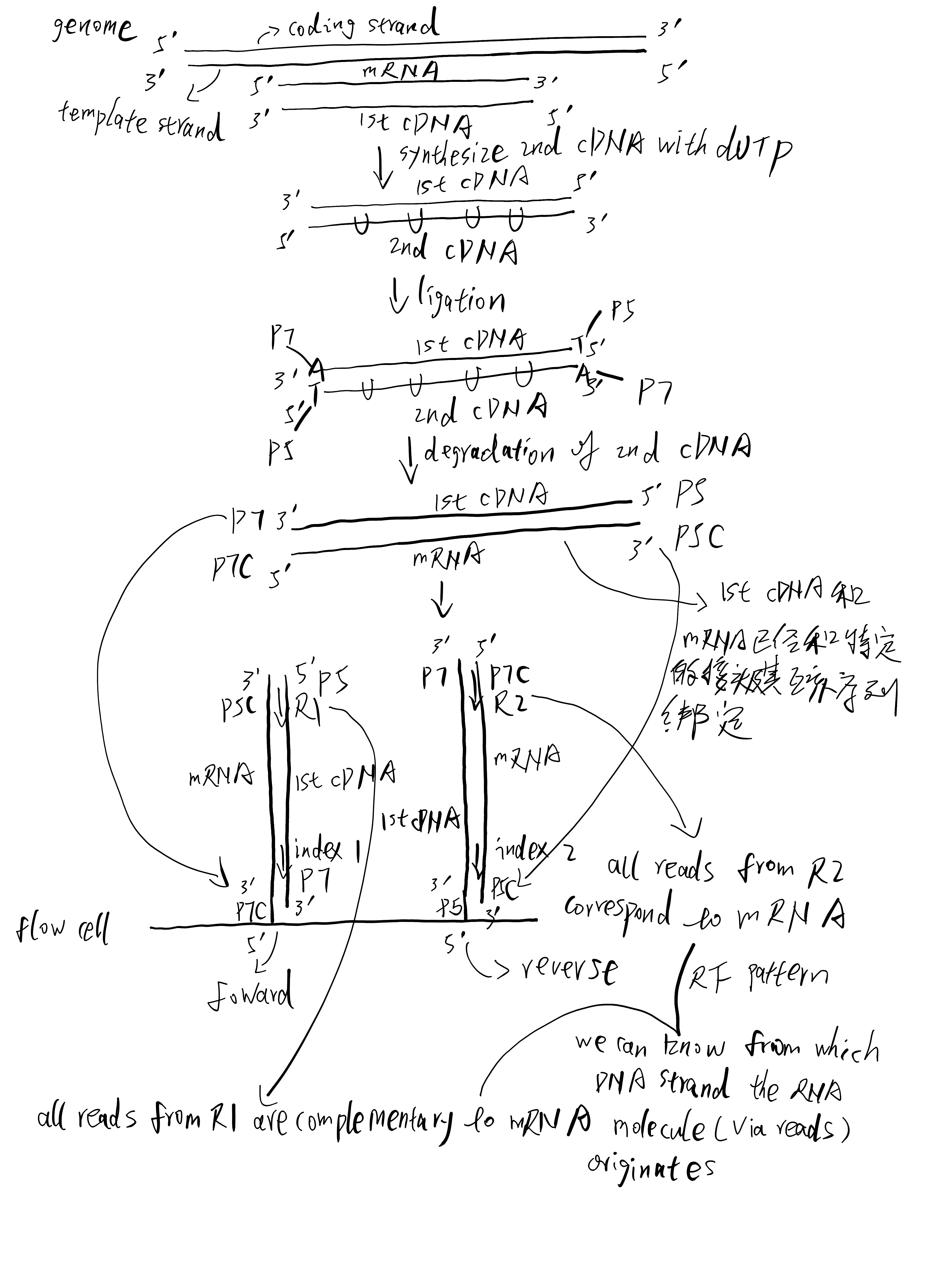
- Via modifications of ds cDNA synthesis: at present, dUTP-based method is most popular.
2 cDNA synthesis method based on dUTP replacement
Next, we’ll see why dUTP-based method can be used for strand-specific RNA-Seq. Now let’s review the dUTP-based protocol step by step:
Purify RNAs with poly-A tails
DNase treatment to reduce DNA contamination
RNA fragmentation
RNA fragmentation can lead to significantly more even reads distribution along the transcript, while this method is sensitive to rRNA contamination.
In ds cDNA shearing scheme, RNA secondary structures distort locally the cDNA synthesis, which leads to non-uniform reads distribution along the RNA, but this will significantly eliminate the influence of rRNA contamination.
Random hexamer-primed first strand synthesis (poly-T-based primer is also commonly used)
Removal of dNTPs
Second strand synthesis with dUTP instead of dTTP
Standard sequencing library preparation procedures
End repair (polishing)
A-tailing
Adaptor ligation
Gel size selection
- UDG treatment to degrade the second strand cDNA with the incorparation of dUTP
Due to the specific ligation direction introduced in the previous step and the removal of the 2nd strand cDNA, we can ensure that reads from R1 (R2) all correspond to RNA molecule or are complementary to it.
Library QC
Library amplification
Sequencing
*Note: Illunima flow cell structure: flow cell \(\longrightarrow\) each flow cell contains a number of lanes \(\longrightarrow\) each lane contains a number of swaths \(\longrightarrow\) each swath contains a number of tiles.
dUTP scheme of ssRNA-Seq: